From forming muscle fibers to protecting us from disease, proteins play an essential role in almost all biological processes in humans and other life forms alike. There has been extraordinary progress in recent years toward better understanding protein structures using deep learning, enabling the accurate prediction of protein structures from their amino acid sequences. However, predicting a single protein structure from its amino acid sequence is like looking at a single frame of a movie—it offers only a snapshot of a highly flexible molecule. Biomolecular Emulator-1 (BioEmu-1) is a deep-learning model that provides scientists with a glimpse into the rich world of different structures each protein can adopt, or structural ensembles, bringing us a step closer to understanding how proteins work. A deeper understanding of proteins enables us to design more effective drugs, as many medications work by influencing protein structures to boost their function or prevent them from causing harm.
One way to model different protein structures is through molecular dynamics (MD) simulations. These tools simulate how proteins move and deform over time and are widely used in academia and industry. However, in order to simulate functionally important changes in structure, MD simulations must be run for a long time. This is a computationally demanding task and significant effort has been put into accelerating simulations, going as far as designing custom computer architectures (opens in new tab). Yet, even with these improvements, many proteins remain beyond what is currently possible to simulate and would require simulation times of years or even decades.
Enter BioEmu-1 (opens in new tab)—a deep learning model that can generate thousands of protein structures per hour on a single graphics processing unit. Today, we are making BioEmu-1 open-source (opens in new tab), following our preprint (opens in new tab) from last December, to empower protein scientists in studying structural ensembles with our model. It provides orders of magnitude greater computational efficiency compared to classical MD simulations, thereby opening the door to insights that have, until now, been out of reach. BioEmu-1 is featured in Azure AI Foundry Labs (opens in new tab), a hub for developers, startups, and enterprises to explore groundbreaking innovations from research at Microsoft.
Microsoft research podcast
What’s Your Story: Lex Story
Model maker and fabricator Lex Story helps bring research to life through prototyping. He discusses his take on failure; the encouragement and advice that has supported his pursuit of art and science; and the sabbatical that might inspire his next career move.
We have enabled this by training BioEmu-1 on three types of data sets: (1) AlphaFold Database (AFDB) (opens in new tab) structures (2) an extensive MD simulation dataset, and (3) an experimental protein folding stability dataset (opens in new tab). Training BioEmu-1 on the AFDB structures is like mapping distinct islands in a vast ocean of possible structures. When preparing this dataset, we clustered similar protein sequences so that BioEmu-1 can recognize that a protein sequence maps to multiple distinct structures. The MD simulation dataset helps BioEmu-1 predict physically plausible structural changes around these islands, mapping out the plethora of possible structures that a single protein can adopt. Finally, through fine-tuning on the protein folding stability dataset, BioEmu-1 learns to sample folded and unfolded structures with the right probabilities.
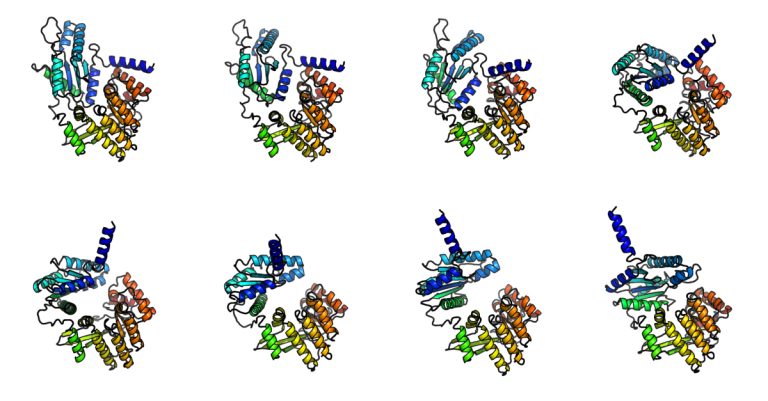
Combining these advances, BioEmu-1 successfully generalizes to unseen protein sequences and predicts multiple structures. In Figure 1, we show that BioEmu-1can predict structures of the LapD protein (opens in new tab) from Vibrio cholerae bacteria, which causes cholera. BioEmu-1 predicts structures of LapD when it is bound and unbound with c-di-GMP molecules, both of which are experimentally known but not in the training set. Furthermore, our model offers a view on intermediate structures, which have never been experimentally observed, providing viable hypotheses about how this protein functions. Insights into how proteins function pave the way for further advancements in areas like drug development.

Moreover, BioEmu-1 reproduces MD equilibrium distributions accurately with a tiny fraction of the computational cost. In Figure 2, we compare 2D projections of the structural distribution of D. E. Shaw research (DESRES) simulation of Protein G (opens in new tab) and samples from BioEmu-1. BioEmu-1 reproduces the MD distribution accurately, while requiring 10,000-100,000 times fewer GPU hours.

Furthermore, BioEmu-1 accurately predicts protein stability, which we measure by computing the folding free energies—a way to quantify the ratio between the folded and unfolded states of a protein. Protein stability is an important factor when designing proteins, e.g., for therapeutic purposes. Figure 3 shows the folding free energies predicted by BioEmu-1, obtained by sampling protein structures and counting folded versus unfolded protein structures, compared against experimental folding free energy measurements. We see that even on sequences that BioEmu-1 has never seen during training, the predicted free energy values correlate well with experimental values.
Professor Martin Steinegger (opens in new tab) of Seoul National University, who was not part of the study, says “With highly accurate structure prediction, protein dynamics is the next frontier in discovery. BioEmu marks a significant step in this direction by enabling blazing-fast sampling of the free-energy landscape of proteins through generative deep learning.”
We believe that BioEmu-1 is a first step toward generating the full ensemble of structures that a protein can take. In these early days, we are also aware of its limitations. With this open-source release, we hope scientists will start experimenting with BioEmu-1, helping us carve out its potentials and shortcomings so we can improve it in the future. We are looking forward to hearing how it performs on various proteins you care about.
Acknowledgements
BioEmu-1 is the result of highly collaborative team effort at Microsoft Research AI for Science. The full authors: Sarah Lewis, Tim Hempel, José Jiménez-Luna, Michael Gastegger, Yu Xie, Andrew Y. K. Foong, Victor García Satorras, Osama Abdin, Bastiaan S. Veeling, Iryna Zaporozhets, Yaoyi Chen, Soojung Yang, Arne Schneuing, Jigyasa Nigam, Federico Barbero, Vincent Stimper, Andrew Campbell, Jason Yim, Marten Lienen, Yu Shi, Shuxin Zheng, Hannes Schulz, Usman Munir, Ryota Tomioka, Cecilia Clementi, Frank Noé